Speeding up genetic diagnosis of Huntington’s disease
Elongated segments of DNA cause Huntington’s disease and certain other disorders of the brain. Researchers funded by the SNSF have developed a method to determine the length of the mutated genes quickly and easily.
People with Huntington’s disease suffer from jerky body movements and decreasing mental abilities. The condition usually leads to death 15–20 years after diagnosis. The cause of the disease is a region in the Huntingtin gene that is longer than in healthy people. The mutation causes the destruction of brain cells.
Five-minute measurement
Determining the length of this gene involves tedious laboratory work and currently takes more than five hours. The team of Vincent Dion, holder of an SNSF professorship at the University of Lausanne, together with collaborators from Toulouse, have developed a reliable method for measuring the length of the responsible DNA region which produces a result within only five minutes (*). The whole diagnosis is thus speeded up more than three times.
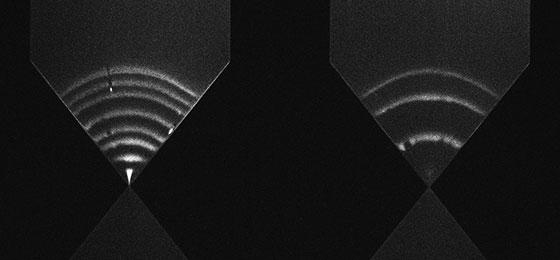
For the analysis, the team extracts the DNA from blood cells, amplifies the concerned region and determines its size with a newly developed chip. The chip holds two small, funnel-shaped chambers a fraction of a millimetre wide. Voltage and pressure are applied to these chambers so as to separate the electrically charged DNA fragments according to their size. The smaller fragments are pushed down the funnel much more than the large ones. By adding a fluorescent dye, the researchers can easily detect the exact position of the fragments under a microscope and deduce their length.
The variable length of the DNA fragments is caused by a repetition of three nucleotides of the genetic code (CAG) – typical for trinucleotide diseases like Huntington’s. The mutation leads to destructive changes in the encoded protein that are currently not fully understood, but the protein produced by the mutated gene is known to be toxic to brain cells. Healthy people have 35 or less of these repeats, whereas most patients have 40 or more. Knowing the exact size is important for forecasting and managing the incurable disease. “Our method is more sensitive and faster than the current methods”, says Vincent Dion.
The project was a collaboration with the group of Aurélien Bancaud from the Laboratoire d'analyse et d'architecture des systèmes in Toulouse, who developed and patented the device. It is licensed out to the company Picometrics Technologies, which has developed the device under the name µLAS.
Cutting out the bad repeats
Huntington’s disease is only one of over twenty known trinucleotide diseases. Others are spinocerebellar ataxia, fragile X syndrome, myotonic dystrophy and Friedreich's ataxia. Currently there is no treatment available for these hereditary diseases. A certain amount of hope is provided by Vincent Dion, who recently developed a method for shortening the fragments with a CRISPR-Cas-based approach (**). “It is, however, still a long way from this proof of concept in cell cultures to a potential medical application”, says Vincent Dion.
(*) R. Malbec et al: µLAS: Sizing of expanded trinucleotide repeats with femtomolar sensitivity in less than 5 minutes. Scientific Reports (2018). DOI:10.1038/s41598-
(**) C. Cinesi et al.: Contracting CAG/CTG repeats using the CRISPR-Cas9 nickase. Nature Communications (2016). DOI:10.1038/ncomms13272
Contact
Promoting young researchers
SNSF Eccellenza Grants allow tenure-track assistant professors to select a new research team and lead an ambitious scientific project. SNSF Eccellenza Professorial Fellowships cover the salaries of assistant professors as well as their project costs. The new scheme replaces the SNSF professorship grant, which has supported 691 researchers since its launch in 2000 – with great success: around 80% of grantees went on to obtain a professorship at a higher education institution in Switzerland or abroad.
Links
- Download picture: (JPEG)Two less than a millimetre wide funnels hold DNA samples of different sizes that fluoresce under laser light. Fragments with known sizes on the left. Test fragments on the right. The shorter they are, the closer the move to the bottom of the funnel. © R. Malbec et al 2017.
- Project in the SNSF research database P3
- Twitter SNSF